OUR VISION
We are mapping the regulatory wiring of the genome to understand the genetic basis of heart diseases.
These maps will enable understanding why heart diseases occur, and designing targeted genome therapies to cure them.
OUR APPROACH
We invent new single-cell methods combining genomics, biochemistry, and molecular biology.
We dissect molecular mechanisms of enhancer-gene communication and gene regulatory networks.
We build computational models to map genome regulation in all cell types in the heart.
We connect human genetic variants to biological mechanisms of disease by applying these tools in cellular and animal models.
We design genome editing therapies to manipulate gene expression to treat heart diseases.
CURRENT RESEARCH
CRISPR tools for understanding genome regulation
We are developing a new suite of tools — combining CRISPR with single-cell readouts — to dissect the functions of variants, regulatory elements, and genes. We use massively parallel genetic screens to enable learning rules of genome biology that generalize across different regions of the genome and across cell types. Together with systematic approaches to observe genomic processes — including chromatin state, RNA expression, and genome structure — the ability to systematically perturb these same processes is transforming genome biology.
Directions in the lab include:
Can we use CRISPR to perturb thousands of enhancers in the genome and precisely quantify their effects on nearby genes at single-cell resolution?
Can we edit thousands of single nucleotides to characterize how human disease variants affect enhancer function and gene expression?
Can we perturb thousands of genes in key cell types in vitro or in vivo to build a comprehensive catalog of gene programs for heart diseases?
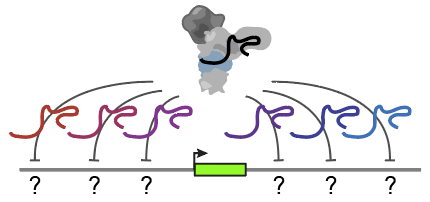
Mapping the regulatory wiring of the genome
Our genomes encode millions of enhancers that tune gene expression in the thousands of cell types in the body. Yet, it has been unclear which enhancers regulate which genes — a massive and complex network that rewires in each cell type. We recently developed an Activity-by-Contact (ABC) model that explains much of this complexity, providing a new strategy to generate comprehensive maps of enhancer-gene connections.
Key questions in the lab include:
How do enhancer-gene connections rewire across cell types, cell states, and dynamic cellular trajectories? Can we directly map these connections using high-throughput CRISPR screens?
Can we refine the ABC model to predict these connections in any cell type and state based on single-cell data?
Can we identify new categories of elements that do not follow the ABC rule, and dissect their mechanisms?
Can we build a complete map of all of the regulatory elements that contribute to heart development?
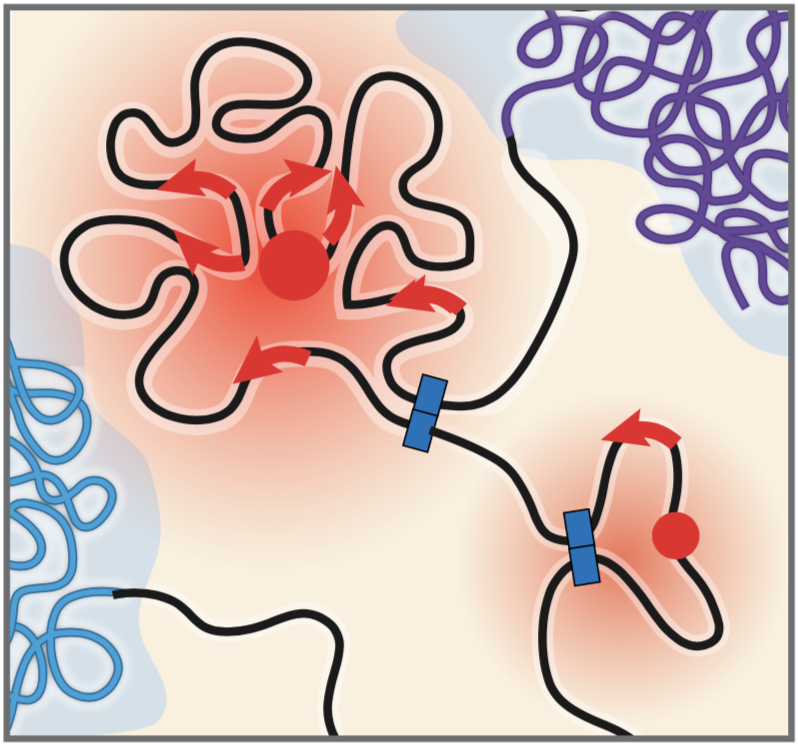
Mechanisms of enhancer-gene communication
We aim to understand the how enhancer-promoter regulation is governed by the complex interplay of transcription factors, 3D chromosome conformation, and noncoding RNAs. Our recent work has revealed a new Activity-by-Contact model for how enhancers regulate specific genes and identified several classes of gene promoters that differ in their responses — yet we still do not understand the precise molecular mechanisms that underlie enhancer Activity and Contact, or how these factors integrate to regulate gene expression.
Key questions in the lab include:
How does 3D Contact quantitatively tune enhancer function?
How does enhancer Activity depend on the synergistic function of multiple enhancers in a locus?
What regulatory proteins mediate the communication between specific classes of enhancers and promoters?
We are combining methods in molecular biology and biochemistry with high-throughput synthetic biology and CRISPR engineering to test hypotheses about enhancer Activity and Contact. And, we are incorporating these insights into our ABC Model to more accurately map the regulatory wiring of the genome.
Connecting Variants to Functions to understand human disease
We are developing new strategies to connect human disease variants to their molecular and cellular functions — for thousands of variants in parallel. Using the ABC model, we have constructed genome-wide maps of enhancer regulation to link enhancers to target genes, and are working on adaptions to single-cell datasets. Using large-scale Perturb-seq, we have constructed maps of gene programs and charted the convergence of disease genes onto particular pathways. To demonstrate these approaches, we study an array of common and rare genetic diseases — including coronary artery disease, congenital heart disease, and other vascular traits. We aim to identify new genes, cell types, and pathways that will guide the development of new treatments for heart diseases.
Key questions in the lab include:
What genes and pathways in which vascular cells control risk for heart diseases and quantitative traits?
Can we systematically connect noncoding variants to their target genes, cell types, and pathways using regulatory maps of enhancer-gene connections and high-content CRISPR screens?
Can we develop new computational tools to combine maps of gene regulation and cellular programs to identify causal genes at GWAS loci?
Can we develop new classes of therapeutics for vascular diseases by editing the regulatory sequences in our genome?